The polymerase chain reaction (PCR) is a laboratory technique for DNA replication that allows a “target” DNA sequence to be selectively amplified. PCR can use the smallest sample of the DNA to be cloned and amplify it to millions of copies in just a few hours.
Discovered in 1985 by Kerry Mullis, PCR has become both and essential and routine tool in most biological laboratories.
Principle of PCR
The PCR involves the primer mediated enzymatic amplification of DNA. PCR is based on using the ability of DNA polymerase to synthesize new strand of DNA complementary to the offered template strand.
Primer is needed because DNA polymerase can add a nucleotide only onto a preexisting 3′-OH group to add the first nucleotide. DNA polymerase then elongate its 3 end by adding more nucleotides to generate an extended region of double stranded DNA.
Components of PCR
The PCR reaction requires the following components:
- DNA Template : The double stranded DNA (dsDNA) of interest, separated from the sample.
- DNA Polymerase : Usually a thermostable Taq polymerase that does not rapidly denature at high temperatures (98°), and can function at a temperature optimum of about 70°C.
- Oligonucleotide primers : Short pieces of single stranded DNA (often 20-30 base pairs) which are complementary to the 3’ ends of the sense and anti-sense strands of the target sequence.
- Deoxynucleotide triphosphates : Single units of the bases A, T, G, and C (dATP, dTTP, dGTP, dCTP) provide the energy for polymerization and the building blocks for DNA synthesis.
- Buffer system : Includes magnesium and potassium to provide the optimal conditions for DNA denaturation and renaturation; also important for polymerase activity, stability and fidelity.
Procedure of PCR
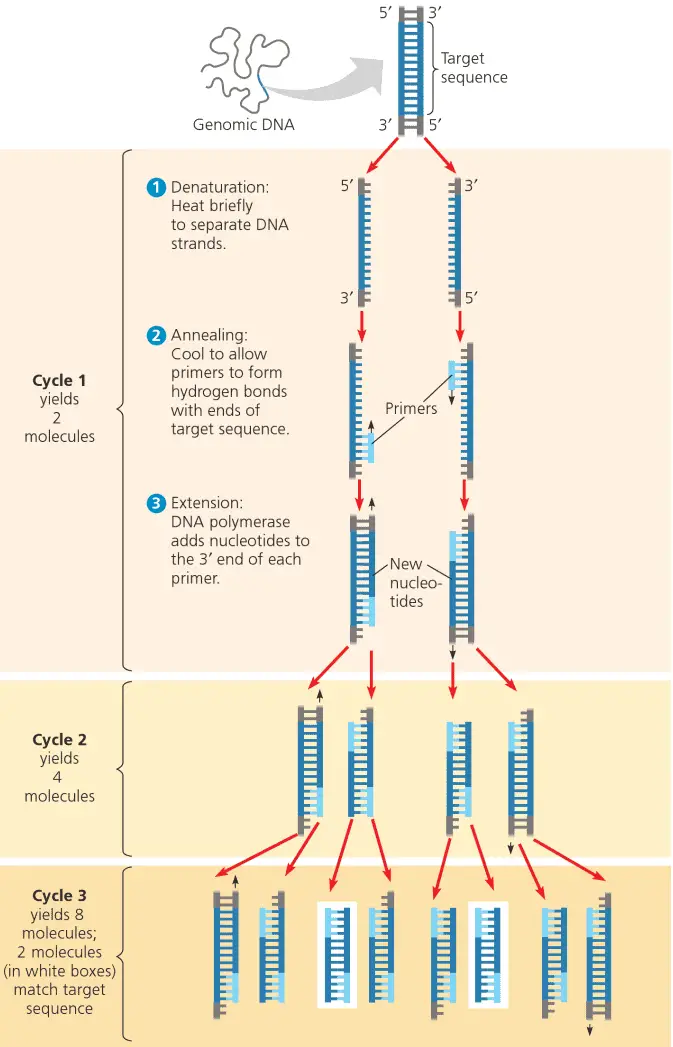
All the PCR components are mixed together and are taken through series of 3 major cyclic reactions conducted in an automated, self-contained thermocycler machine.
- Denaturation :
This step involves heating the reaction mixture to 94°C for 15-30 seconds. During this, the double stranded DNA is denatured to single strands due to breakage in weak hydrogen bonds. - Annealing :
The reaction temperature is rapidly lowered to 54-60°C for 20-40 seconds. This allows the primers to bind (anneal) to their complementary sequence in the template DNA. - Elongation :
Also known at extension, this step usually occurs at 72-80°C (most commonly 72°C). In this step, the polymerase enzyme sequentially adds bases to the 3′ each primer, extending the DNA sequence in the 5′ to 3′ direction. Under optimal conditions, DNA polymerase will add about 1,000 bp/minute.
With one cycle, a single segment of double-stranded DNA template is amplified into two separate pieces of double-stranded DNA.
These two pieces are then available for amplification in the next cycle. As the cycles are repeated, more and more copies are generated and the number of copies of the template is increased exponentially.
Types of PCR
In addition to the amplification of a target DNA sequence by the typical PCR procedures already described, several specialised types of PCR have been developed for specific applications.
- Real-time PCR
- Quantitative real time PCR (Q-RT PCR)
- Reverse Transcriptase PCR (RT-PCR)
- Multiplex PCR
- Nested PCR
- Long-range PCR
- Single-cell PCR
- Fast-cycling PCR
- Methylation-specific PCR (MSP)
- Hot start PCR
- High-fidelity PCR
- In situ PCR
- Variable Number of Tandem Repeats (VNTR) PCR
- Asymmetric PCR
- Repetitive sequence-based PCR
- Overlap extension PCR
- Assemble PCR
- Intersequence-specific PCR(ISSR)
- Ligation-mediated PCR
- Methylation –specifin PCR
- Miniprimer PCR
- Solid phase PCR
- Touch down PCR, etc
Applications of PCR
Some common applications of PCR in various fields can be explained in following categories.
Medical Applications:
- Genetic testing for presence of genetic disease mutations. Eg: hemoglobinopathies, cystic fibrosis, other inborn errors of metabolism
- Detection of disease causing genes in suspected parents who act as carriers.
- Study of alteration to oncogenes may help in customization of therapy
- Can also be used as part of a sensitive test for tissue typing, vital to organ transplantation
- Helps to monitor the gene in gene therapy
Infectious disease Applications:
- Analyzing clinical specimens for the presence of infectious agents, including HIV, hepatitis, malaria, tuberulosis etc.
- Detection of new virulent subtypes of organism that is responsible for epidemics.
Forensic Applications:
- Can be used as a tool in genetic fingerprinting. This technology can identify any one person from millions of others in case of : crime scence, rule out suspects during police investigation, paternity testing even in case of avaibility of very small amount of specimens ( stains of blood, semen, hair etc)
Research and Molecular Genetics:
- In genomic studies: PCR helps to compare the genomes of two organisms and identify the difference between them.
- In phylogenetic analysis. Minute quantities of DNA from any source such a fossilized material, hair, bones, mummified tissues.
- In study of gene expression analysis, PCR based mutagenesis
- In Human genome project for aim to complete mapping and understanding of all genes of human beings.
Frequently Asked Questions
Q 1. What is the purpose of a polymerase chain reaction?
Q 2. What happens in a polymerase chain reaction?
Q 3. What is needed for PCR?
1. DNA sample
2. ddNTPs (free nucleotides)
3. DNA primers
4. DNA polymerase
Q 4. How is the PCR used to diagnose?
– It is used to find out the viral load of HIV in patients suffering from AIDS.
– It is helpful in determining the number of cancerous cells that are remaining in a cancer patient undergoing treatment.
Q 5. Is real-time PCR quantitative?
Q 6. What is the end result of PCR?
Q 7. What happens at 72 degrees in PCR?
Q 8. How many types of PCR are there?
Real-time PCR/Quantitative PCR/qPCR – It uses a fluorescent dye to tag the molecules of DNA. it is used to detect and quantify PCR products in real-time.
Multiplex PCR – It multiplies multiple fragments in a single sample of DNA using a number of primers.
reverse-transcriptase – The purpose is to create complementary DNA by means of reverse transcribing RNA to DNA with the help of reverse transcriptase.
Hot start PCR – Heat is used to denature antibodies that are used for Taq polymerase inactivation.
Nested PCR – Once the initial PCR cycle is done, another PCR is done but this time with the use of a new primer nested within the original primer. Thus, the term nested PCR. The reason for doing so is to reduce the risk of unwanted products.
Assembly PCR – Overlapping primers are used to amplify longer fragments of DNA.
Long-range PCR – A longer range of DNA is formed with the help of a polymerase mixture.
In situ PCR – It is a type of PCR that takes place in the cells or fixed tissue on a slide.
Asymmetric PCR – A single stand of target DNA is amplified.
Q 9. How accurate is a polymerase chain reaction?
Q 10. What diseases can PCR detect?
1. Hepatitis
2. HIV
3. Human papillomavirus (causes genital warts and cervical cancer)
4. Malaria
5. Anthrax
6. Epstein-Barr virus in people with glandular fever
Q 11. What do PCR primers do?
Q 12. Why is PCR important?
Q 13. What enzyme is used for PCR?
Q 14. What are the 4 steps of PCR?
1. The first step is denaturation using heat.
2. The second step is annealing the primer to a specific target sequence of DNA.
Extension
3. End of the first cycle.

Sir, V.good topic plz continue it.
Thank you very much for the this topic
sir very happy this notes thank you..
Thank you
its very imp.continue
thanks..
Well Done
thanx so much….really helpful
Interesting explanation
Its very easy to understand
A very lucid and explicit note for easy understanding. Looking forward to reading more of it kind.
its wht i need right now…for my study…thnk you verry much
thank u very much, the info is very brief and helpful.thanks a lot
Straight to d point, rily helpful, tnx sir.
Thanks for the information